The microbial-metabolic nexus in colon cancer
Cutting-edge colon adenocarcinoma research reveals how fatty acid metabolism, intratumoral microbiota, and the tumor microenvironment shape patient outcomes, paving the way for AI-driven diagnostic innovations in cancer care.
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
About this article
Author
Intratumoral dynamics in colon cancer are complex, with microbiota, (sidenote: Fatty Acid Metabolism (FAM) The cellular process involved in breaking down and synthesizing fats, influencing tumor growth and progression. ) , and the (sidenote: Tumor Microenvironment (TME) The surrounding environment of a tumor, including immune cells, blood vessels, and signaling molecules that impact cancer development. ) all playing a role. A recent study 1 delved into this "unresolved trinity" in (sidenote: Colon Adenocarcinoma (COAD) A type of colon cancer originating in the glandular cells of the colon. ) . By leveraging a substantial cohort of patient data from The Cancer Genome Atlas (TCGA) and employing sophisticated bioinformatic and pathological image analysis techniques, the researchers aimed to uncover novel diagnostic and therapeutic avenues for this aggressive cancer.
Microbial-metabolic connections
The research team initiated their investigation by analysing comprehensive data from 420 patients diagnosed with COAD. A key methodological step involved categorising these patients into two distinct subgroups:
- those exhibiting high fatty acid metabolism (FAM_high)
- and those with low fatty acid metabolism (FAM_low).
This stratification based on the (sidenote: Gene Set Variation Analysis (GSVA) A computational method used to evaluate the activity of specific gene pathways in patient samples, aiding in diagnostic subtyping. ) score calculated for FAM pathway genes.
Surprisingly, despite the overall microbial alpha diversity (a measure of within-sample diversity) appearing similar between the two groups, deeper analysis of the microbial beta diversity (a measure of between-sample diversity) revealed remarkably distinct bacterial compositions strongly linked to these underlying metabolic profiles.
Specific types of gut-associated bacteria were found to play a role in modulating the tumor environment. Notably, the study identified a panel of specific bacterial genera, including Desulfovibrio, Desulfococcus, Streptococcus, and Mycobacterium, that were significantly enriched within the FAM_high patient group, showing a clear connection between the (sidenote: Intratumoral Microbiota The community of microorganisms present within the tumor, which can affect its behavior and the patient's prognosis. ) and host metabolism. This metabolic stratification also had prognostic significance.
The researchers observed that patients whose tumors exhibited a low FAM signature (FAM_low group) experienced significantly better overall survival (OS) compared to their FAM_high counterparts. The identification of four specific genes (ADIPOR2, HAO2, ALAD, HPGD) whose expression levels were significantly correlated with patient survival, solidifying the critical prognostic role of FAM in COAD.
Metabolic signatures as predictive tools
Beyond diagnosis and prognosis, the study's comprehensive drug sensitivity analysis illuminated substantial and potentially clinically actionable differences in how the two FAM-defined subtypes responded to a broad spectrum of therapeutic agents.
By calculating the IC50 values (the concentration of a drug required to inhibit 50% of cells) for an extensive panel of 195 candidate drugs, the researchers uncovered that the FAM_high and FAM_low groups exhibited significantly different sensitivities to a remarkable 120 of these compounds. For example, the high FAM group showed less sensitivity to drugs like JQ1, suggesting these agents might be less effective in this metabolic context. Such insights could support more personalized cancer treatment strategies.
Imaging insights for diagnosis
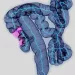
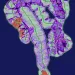
Perhaps most notably, the research found that standard histopathology images can reflect the underlying FAM subtypes. Distinct texture features correlated with FAM scores and microbial signatures, differing significantly between the high and low FAM groups. AI models could soon analyze histology to detect bacterial signatures and tumor metabolism status in real time. This suggests the exciting possibility of developing AI-powered tools to predict metabolic subtypes from routine pathology, offering a cost-effective and accessible diagnostic approach.
This research provides significant insights into the intricate relationship between the gut and tumor environment in COAD progression.. The potential for non-invasive metabolic subtyping through AI-driven image analysis holds particular promise for broad clinical translation. A deeper understanding of how gut-derived bacteria interact with tumor cells will be key to future therapeutic advances.