Microbiota and celiac disease
Overview
By Pr. Elena F. Verdu
Farncombe Family Digestive Health Research Institute, McMaster University Hamilton, Canada
Lay public section
Find here your dedicated section
Sources
This article is based on scientific information
Sections
About this article
Author
Environmental factors are suggested to contribute to celiac disease pathogenesis, an autoimmune disease triggered by the ingestion of gluten. Clinical studies show alterations in the composition of the microbiota in celiac disease patients. Although some consistent findings across studies have been established, no celiac disease microbial signature has been defined. Using reductionist and gnotobiotic* animal models, recent research suggested that bacterial strains from celiac disease patients may have more pathogenic or inflammatory potential. Targeting the microbiota with the use of specific probiotics, shown to modify pathogenic mechanisms critical for celiac disease, could be an attractive therapeutic approach to complement the gluten-free diet.
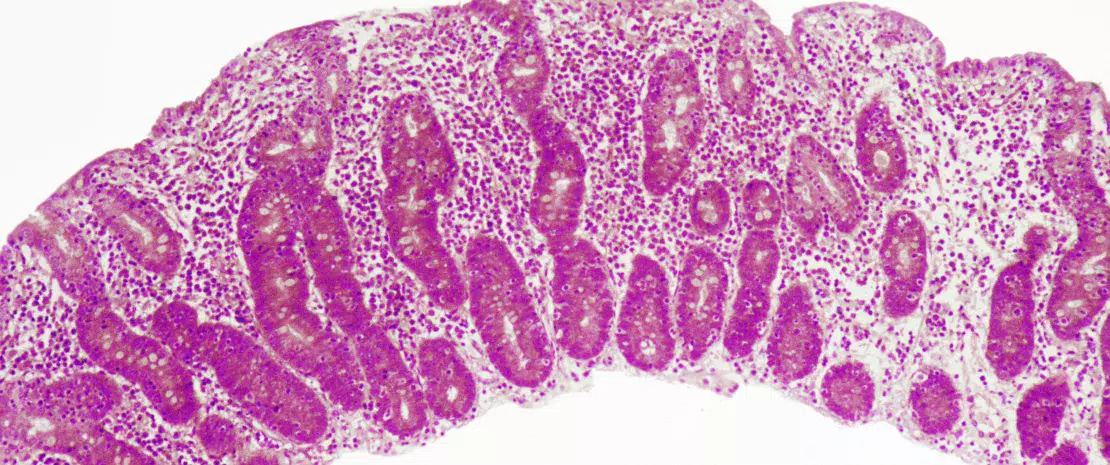
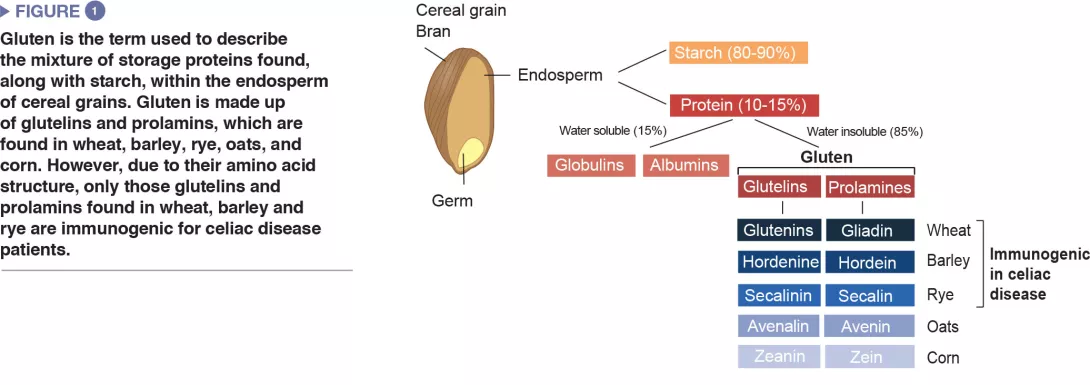
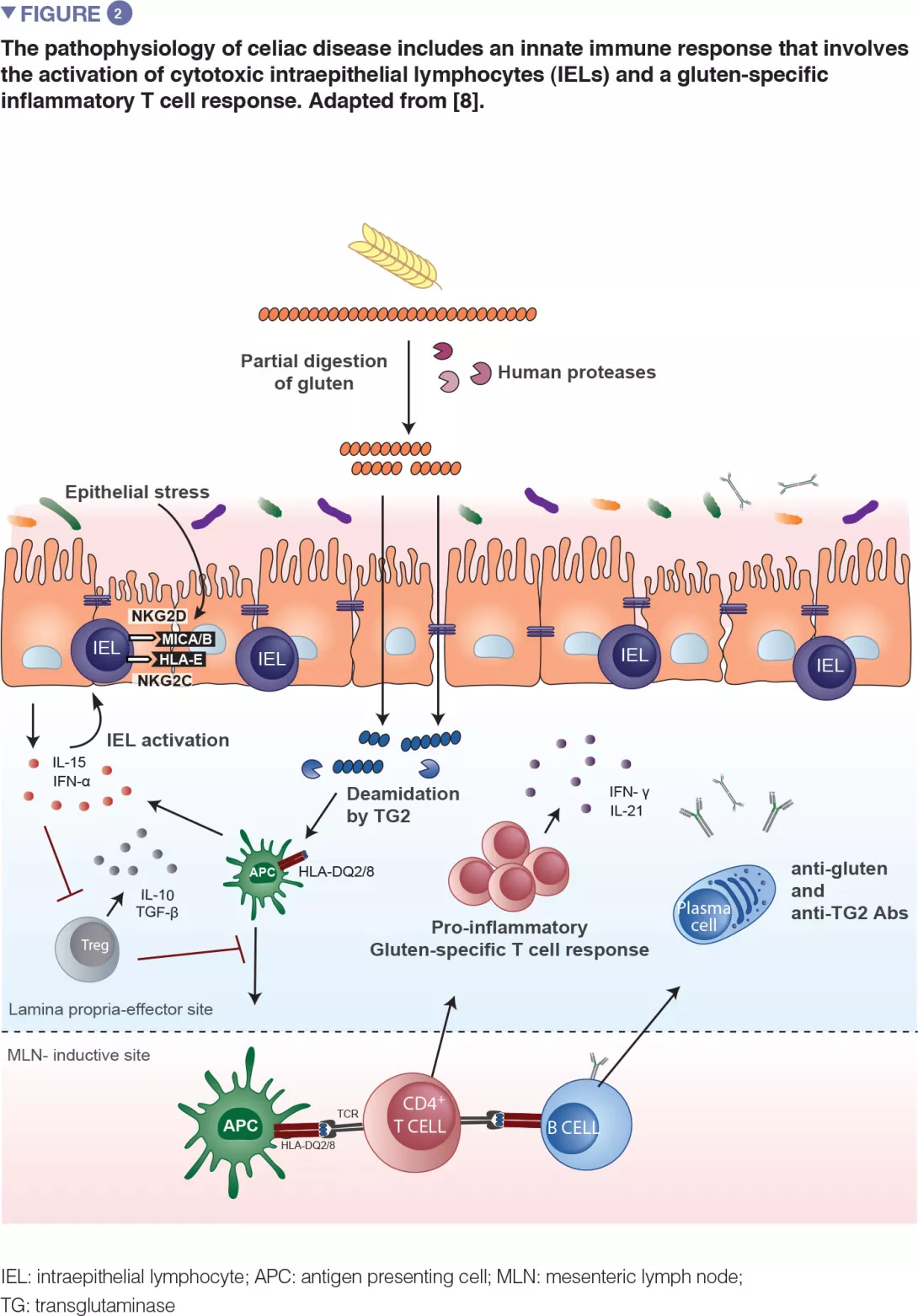
Celiac disease is a common inflammatory and autoimmune reaction that occurs in genetically predisposed individuals after consuming gluten (Figure 1). The characteristic lesion is the destruction of the finger-like projections of small intestinal lining (enteropathy). Clinical manifestations of the disease are varied and include both intestinal or extra-intestinal symptoms. Celiac disease is unique in that it is the only autoimmune disease where the triggering antigen (gluten) is known. The mechanisms that explain HLA genetic risk and the steps triggered by the dietary trigger that ultimately leads to the development of pro-inflammatory gluten specific T-cells and autoantibodies, are well understood (Figure 2). One unsolved question relates to the rapid recent increase in prevalence as well as the fact that celiac disease only develops in a fraction of genetically susceptible individuals, suggesting there must be additional genetic or environmental factors involved in activating the inflammatory cascade. In particular, there has been a growing interest in the role of the microbial factors in celiac disease development [1]. In this review we focus on bacterial alterations and how they could play a role in disease mechanisms as well as how they constitute potential therapeutic targets for celiac disease.
Correlation of dysbiosis and celiac disease: lessons from clinical studies
One of the first studies to suggest a microbial contribution to celiac disease described the presence of rod-shaped bacteria in duodenal biopsies of children born during a celiac disease “epidemic” in Sweden. These bacteria were not observed in children without celiac disease, or in children born following the epidemic and it was thought that their presence may have contributed to the increase in the disease incidence observed in Sweden.[2] However, the mechanisms underlying this association remain unknown.
A number of studies have since been published that analyzed the fecal and small intestinal microbiota composition in celiac disease patients compared to healthy controls. Some relatively consistent findings across studies include increases in proportions of Bacteroides and members of Proteobacteria, and decreases in Lactobacillus and Bifidobacteria in celiac patients compared to controls.[1] Increased abundance of Proteobacteria was also found in patients who suffered from persistent symptoms, despite adhering to a gluten-free diet.[3] More recently, children at a high genetic risk for developing celiac disease were shown to have a different microbiota composition compared to children who were at a low genetic risk.[4-6] Finally, at-risk children who went on to develop celiac disease were suggested to have higher basal microbial diversity that did not further diversify over time, suggestive of a “premature maturation” of the gut microbiota.[7] While the results suggest that an altered early trajectory of the microbiota could predispose to celiac disease, larger trials with increased sample sizes are needed to confirm the findings. Despite the findings of an altered composition of the microbiota in celiac disease or in at-risk children, no celiac “microbial signature” has been established. Differences in the locations of study population, status of control subjects, fecal vs. small intestinal samples, and methodology may contribute to inconsistencies between studies. Inconsistent findings have also been reported regarding associations between events that can alter the development of the microbiota and celiac disease develop- ment. While early studies suggested anti- biotic use and delivery by C-section could increase celiac disease risk, more recent larger clinical studies did not confirm these associations.[8]
The long-term follow-up of at-risk infants may provide insight into the factors that may contribute to disease onset. Despite no evidence for causation, these clinical associations have stimulated the study of basic causative mechanisms in reductionist systems and animal models.
Mechanisms by which microbiota alterations can influence celiac disease: lessons from basic research
How microbes could contribute to the pathogenesis or development of celiac disease can be better understood by studying the function of the microbial community in celiac disease patients vs. healthy subjects. Isolation of bacteria from the human small intestine allows for translation into reductionist models. For instance, strains of Enterobacteriaceae isolated from celiac patients were more virulent than those isolated from healthy controls.[9] Moreover, E. coli strains isolated from celiac children had more in vitro pro-inflammatory capacity compared to strains of Bifidobacterium that were isolated from control children.[10] Microbiota “humanized” mouse models of germ-free mice add a layer of complexity and allow in vivo comparison of phenotypes induced by a microbiota of interest. Moreover, these mice can express features of the human immune system (such as MHC-class II expression) that are critical for celiac disease development. Transgenic mice that express the human celiac risk gene, HLA-DQ8, were protected from gluten-induced pathology when they were minimally colonized with a microbiota that was free from pathogens or opportunistic bacteria. However, if an adherent strain of E. coli, isolated from the celiac gut, was added to the protective bacteria, mice developed gluten-induced pathology. Similarly, treatment of mice harbouring a diverse murine microbiota with the antibiotic vancomycin led to an increase in Proteobacteria, including E. coli, and a worsening of gluten-induced pathology.[11]
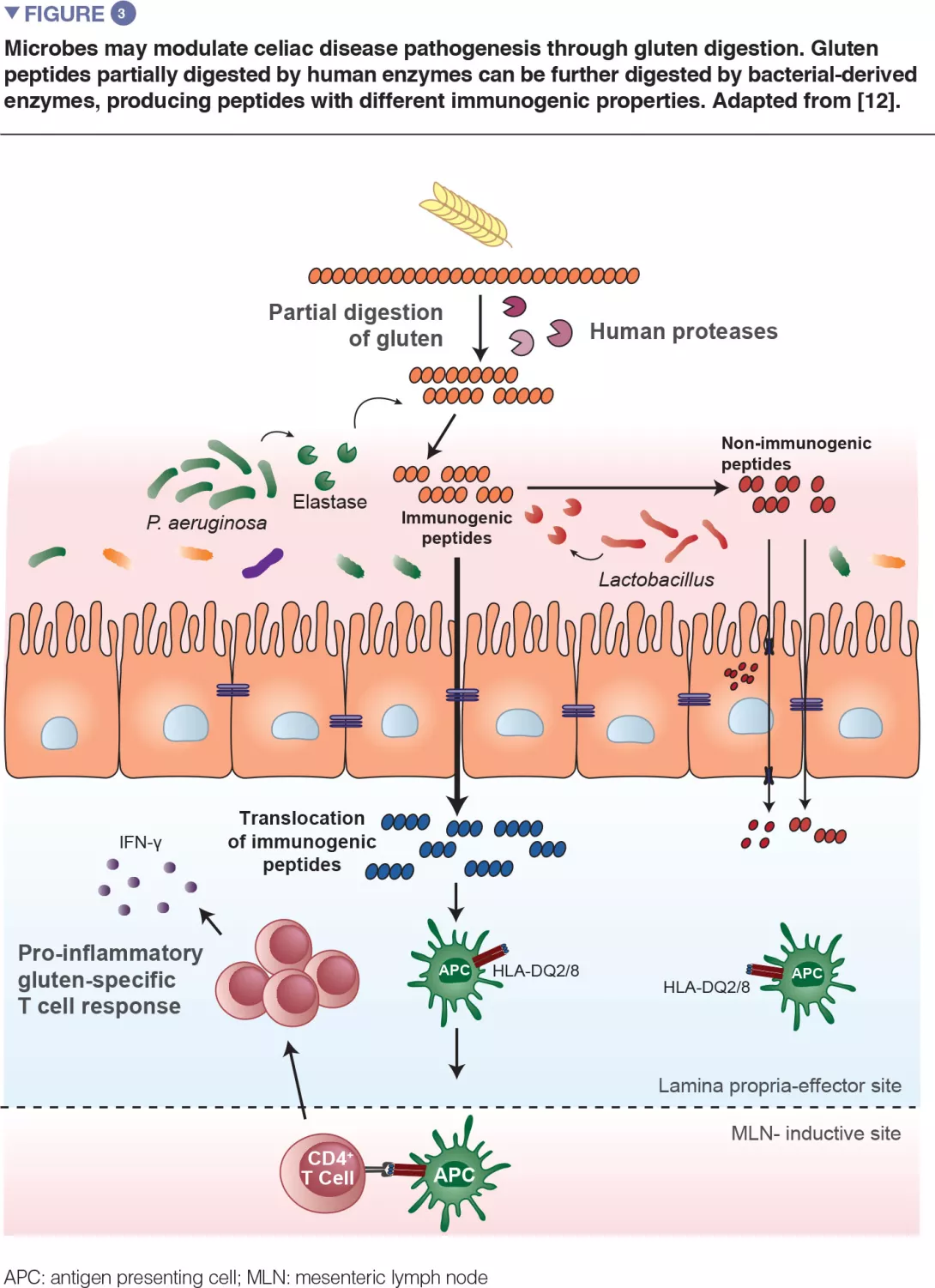
Recent translational work in mice expanded on the observation that bacteria are capable of degrading gluten (Figure 3). The study used gnotobiotic mice that were colonized with either opportunistic pathogens, such as Pseudomonas aeruginosa, or with commensals, such as Lactobacillus. The authors showed that different bacteria can degrade gluten in vivo, but the protein fragments they produce are distinct. The study further demonstrated that enzymes from P. aeruginosa, which was isolated from a celiac disease patient, could degrade gluten. This digestion process produced gluten fragments that stimulated an inflammatory immune response in cells isolated from celiac disease patients and were better able to cross the small intestinal barrier, where interaction with immune cells would occur. Several peptides generated by P. aeruginosa digestion that were subsequently digested with lactobacilli, isolated from a healthy subject and a core member of the healthy microbiome, no longer induced inflammatory immune responses in vitro. This study provided a key mechanism where both opportunistic pathogens and commensals may modify the repertoire and immune properties of gluten peptides in the gut, thereby impacting disease susceptibility.[12]
Can microorganisms be used to treat or prevent celiac disease?
A diagnosis of celiac disease means strict, life-long avoidance of gluten-containing foods because exposure to small amounts of gluten can trigger a variety symptoms and enteropathy in affected people. Gluten is used ubiquitously in processed foods, which makes strict adherence difficult, and has prompted the search for alternative or adjuvant therapies. Given the key role for microorganisms in regulating immunity and the association between celiac disease and altered composition and function of the microbiota, the therapeutic potential of several probiotics has been tested. A strain of Bifidobacterium longum, previously shown to have anti-inflammatory effects in vitro,[10-13] was tested in children on a gluten-free diet in a double-blind placebo-controlled trial.
Gluten metabolism by bacteria
-
Gluten is highly resistant to breakdown by host digestive enzymes in the small intestine due to its amino acid structure.
-
This leaves large gluten fragments that are capable of inducing immune responses once they cross the epithelial barrier in genetically susceptible individuals.
-
The gastrointestinal tract harbours bacteria that are able to breakdown gluten, and these bacteria may differ between celiac disease patients and healthy subjects.[17]
Administration of the probiotic led to some immune changes, as well as lower levels of potentially harmful bacteria (B. fragilis). However, no changes in symptoms were observed between children that received the probiotic compared to those that received placebo.[14] Because the probiotic was administered together with the gluten-free diet, it is difficult to discern between changes induced by the dietary restriction or the probiotic. Two other studies tested the effects of a strain of Bifidobacterium infantis. The first randomized double-blind placebo-controlled trial demonstrated that patients receiving the probiotic showed significant improvement of symptoms following the 3-week trial, but no differences were found in intestinal permeability.[15] A follow-up trial tested whether the same probiotic could modulate innate immune responses, which could be responsible for the symptomatic improvement previously observed. Administration of a strain of B. infantis led to a decrease in the number of small intestinal Paneth cells, that paralleled a decrease in antimicrobial peptides. These effects of the probiotic were independent of the gluten free diet.[16] Due to the nature of these few studies including only small groups, there is no evidence to date to recommend any probiotic in celiac disease. Moreover, probiotics consumed by patients with celiac disease need to be rigorously certified gluten-free, and this is not the case for every over the counter preparation. Prior to patient consumption, we need a better understanding of mechanisms of action, and those chosen for further testing should be selected due to their involvement in celiac disease pathways. For example, bacteria that aid in the detoxification of gluten could be selected and used to complement the gluten-free diet. However, to date, no single bacterium tested has shown optimal gluten digestion in vitro. Studies have focused on bacterial strains that produce enzymes capable of degrading gluten, but fungal species such as Aspergillus niger, also produce gluten-degrading enzymes, and rational combinations of fungal and bacterial organisms may offer an attractive avenue of therapeutic research in celiac disease.
Conclusion
The role of the intestinal microbiota in celiac disease has become evident. Expanding on clinical associations, reductionist systems and gnotobiotic animal models have provided evidence that specific microbes can modulate key steps in celiac disease pathogenesis. The continued use of these systems to study specific microbe-host and microbe-gluten interactions as well as larger clinical studies where at-risk individuals are followed over time are critical for understanding how microbes could trigger disease. This can allow for microbe targeted preventative strategies or adjuvant therapies to the gluten-free diet.