Microbiota-derived 3-IAA (indole-3-acetic acid) influences chemotherapy efficacy in pancreatic cancer
COMMENTED ARTICLE - ADULTS’ SECTION
By Pr. Harry Sokol
Gastroenterology and Nutrition Department, Saint-Antoine Hospital, Paris, France
Commentary on the article by Tintelnot et al. Nature 2023 [1]
Pancreatic ductal adenocarcinoma (PDAC) is expected to become the second most deadly cancer by 2040, owing to the high incidence of metastatic disease and limited responses to treatment. Less than half of all patients respond to the primary treatment for PDAC, and genetic alterations alone cannot explain sufficiently the different responses. Diet is an environmental factor that can influence the response to therapies, but its role in PDAC is unclear. Here, using shotgun metagenomic sequencing and metabolomic screening, the authors showed that the microbiota-derived tryptophan metabolite indole-3-acetic acid (3-IAA) is enriched in patients who respond to treatment. Faecal microbiota transplantation, dietary manipulation of tryptophan and oral 3-IAA administration increase the efficacy of chemotherapy in humanised mouse models of PDAC. Using a combination of loss- and gain-of-function experiments, the authors showed that the efficacy of 3-IAA and chemotherapy requires the presence of neutrophil-derived myeloperoxidase (MPO). MPO oxidises 3-IAA which, in combination with chemotherapy induces a downregulation of the reactive oxygen species (ROS)-degrading enzymes glutathione peroxidase 3 and glutathione peroxidase 7. This all results in the accumulation of ROS and the downregulation of autophagy in cancer cells, which compromises their metabolic fitness and, ultimately, their proliferation. In humans, the authors observed a significant correlation between 3-IAA levels and the efficacy of therapy in two independent PDAC cohorts. In summary, the authors identified a microbiota-derived metabolite that has clinical implications in the treatment of PDAC, and provided a motivation for considering nutritional interventions when treating cancer patients.
What do we already know about this subject?
Polychemotherapy, either with 5-fluorouracil (5-FU), irinotecan and oxaliplatin in combination with folinic acid (FOLFIRINOX), or with gemcitabine and nabpaclitaxel (GnP), is considered the standard of care for patients suffering from metastatic PDAC (mPDAC). However, less than half of all patients are responsive to the therapy, and patients who do not respond (NR, non-responder patients) suffer pain and die within a few weeks. Genetic alterations in PDAC poorly explain the differences between patients who respond to therapy (R, responder patients) and NR patients, which leaves environmental factors, including the intestinal microbiota, as the potential mediators of chemotherapy efficacy. Thus, there is an urgent need to identify environmental factors that might explain the differences between R and NR patients to develop new concepts for future therapies. The intestinal microbiota has been shown to induce a response to immunotherapy in patients with melanoma, and can be modulated by the diet 2, 3 . In rare long-term survivor patients with localised PDAC, bacteria can pass from the intestine to the tumour and thus control anti-tumour immune activation. However, most patients suffering from aggressive immunotherapy-resistant mPDAC are treated with polychemotherapy, and it is presently unclear whether and how the microbiota or dietary habits affect the efficacy 4 .
- The intestinal microbiota is different in PDAC patients who are responders and non-responders to chemotherapy
- The tryptophan metabolite 3-IAA, produced by the microbiota, is enriched in responder patients
- 3-IAA increases the efficacy of chemotherapy by boosting ROS production by neutrophils and decreasing autophagy in cancer cells
What are the main insights from this study?
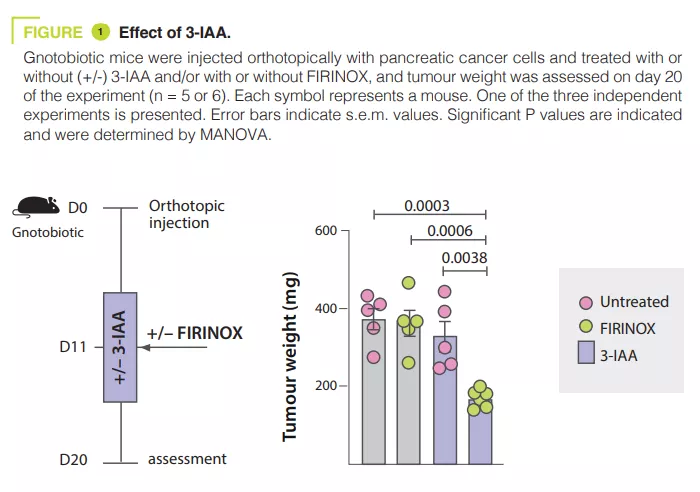
Analysing the microbiota of 30 mPDAC patients revealed differences between R and NR patients. Transferring the microbiota of R patients to mice with pancreatic tumours reduced the size of the tumours after chemotherapy. The tryptophan metabolite 3-IAA was enriched in R patients and mice with R microbiota, potentially contributing to the response to chemotherapy. The administration of 3-IAA increased the efficacy of chemotherapy in mice (figure 1). The analysis of immune cells in mice showed an increase in CD8+ T cells and a reduction in neutrophils in mice with a microbiota associated with a good response to chemotherapy. 3-IAA affected the MPO of neutrophils, thereby reducing their survival. The combination of 3-IAA and chemotherapy reduced the number of neutrophils and inhibited tumour growth, with MPO playing a crucial role. It is suggested that MPO induces the ROS production leading to cell death during chemotherapy. In-vitro experiments have shown that 3-IAA increases ROS levels. This effect was confirmed in vivo and the inhibition of ROS by N-acetylcysteine abolished the efficacy of FIRINOX in mice with high 3-IAA levels. The authors then showed that the effect of 3-IAA was linked to downregulation of autophagy. Finally, high serum concentrations of 3-IAA were correlated to a reduction in neutrophil counts and improved survival in the two cohorts of human patients.
What are the consequences in practice?
The intestinal microbiota has an effect on chemotherapy responses. The mechanisms involved in this study demonstrates the role of microbial metabolites, particularly tryptophan metabolites. Among these, 3-IAA is not only a predictive marker of response to chemotherapy in PDAC, but could also offer an adjuvant therapeutic drug.
CONCLUSION
The intestinal microbiota has an effect on chemotherapy responses in PDAC. One of its metabolites, 3-IAA, is a predictor of a good response to chemotherapy and enhances its effect by inducing an accumulation of ROS and reducing autophagy in cancer cells.
1.Tintelnot J, Xu Y, Lesker TR, et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 2023 ; 615 : 168-74.
2. Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021 ; 371 : 602-9.
3. Spencer CN, McQuade JL, Gopalakrishnan V, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021 ; 374 : 1632-40.
4. Thomas RM, Jobin, C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol 2020 ; 17 : 53-64.