Metagenomics of the faecal virome: cumulative effect of the quantity of gluten and enteroviruses risk of coeliac disease autoimmunity in predisposed children – the TEDDY study
Commented articles - Children's section
By Pr. Emmanuel Mas
Gastroenterology and Nutrition Department, Children’s Hospital, Toulouse, France
Commentary on the original article of Lindfors et al. (Gut 2019) [1]
A higher gluten intake, frequent gastrointestinal infections and adenovirus, enterovirus, rotavirus and reovirus have all been proposed as environmental triggers of coeliac disease (CD). However it is not known whether an interaction exists between the quantity of gluten ingested and exposure to viruses in the development of CD. This study sought to determine whether distinct viral exposures, alone or associated with gluten, increased the risk of CD autoimmunity in genetically predisposed children. It was concluded that frequent exposure to enteroviruses between the ages of 1 and 2 years was indeed associated with increased risk of CD autoimmunity, indicating a cumulative effect of the interaction between enteroviruses and higher gluten intake.
What do we already know about this subject?
Coeliac disease is an autoimmune pathology which occurs in genetically predisposed individuals of genotype HLA DQ2 and/ or DQ8-positive. It is characterised by the presence of villous atrophy and lymphocyte infiltration of the epithelium of the small intestine. Gluten present in the diet induces an autoimmune response directed against tissue transglutaminase. The appearance of anti-transglutaminase antibodies (ATA) indicates the presence of coeliac disease autoimmunity
The rise in the incidence of autoimmune diseases has led to suspect that environmental factors may have a role in their pathogenesis. Observational studies suggest that viral infections could cause a loss of oral tolerance to gluten and the development of coeliac disease.
What are the main insights from this study?
It is a nested case-control study in the TEDDY (The Environmental Determinants of Diabetes in the Young) birth cohort which included 8,676 children before the age of 4 and a half months followed to the age of 15 years. The principal objective of this cohort was to identify the genetic and environmental factors associated with type 1 diabetes and coeliac disease. After forming pairs matched for family history of type 1 diabetes, sex and study inclusion site, 83 pairs (child with predisposition (case) and control) were retained in the final analysis for whom faecal virome data were available after introduction of gluten. Of these pairs, 16 had a family history of type 1 diabetes. During follow-up, 28 of the coeliac disease autoimmunity cases developed coeliac disease.
Stool samples were collected every month from the age of 3 months to 2 years; tests for enterovirus, adenovirus, astrovirus, norovirus, reovirus and rotavirus were performed. Every 3 months, a food questionnaire was used to collect information on breast-feeding and the age of introduction of gluten-containing foods. A 3-day record of food intake enabled a calculation of the quantities of gluten ingested at 6, 9, 12, 18 and 24 months.
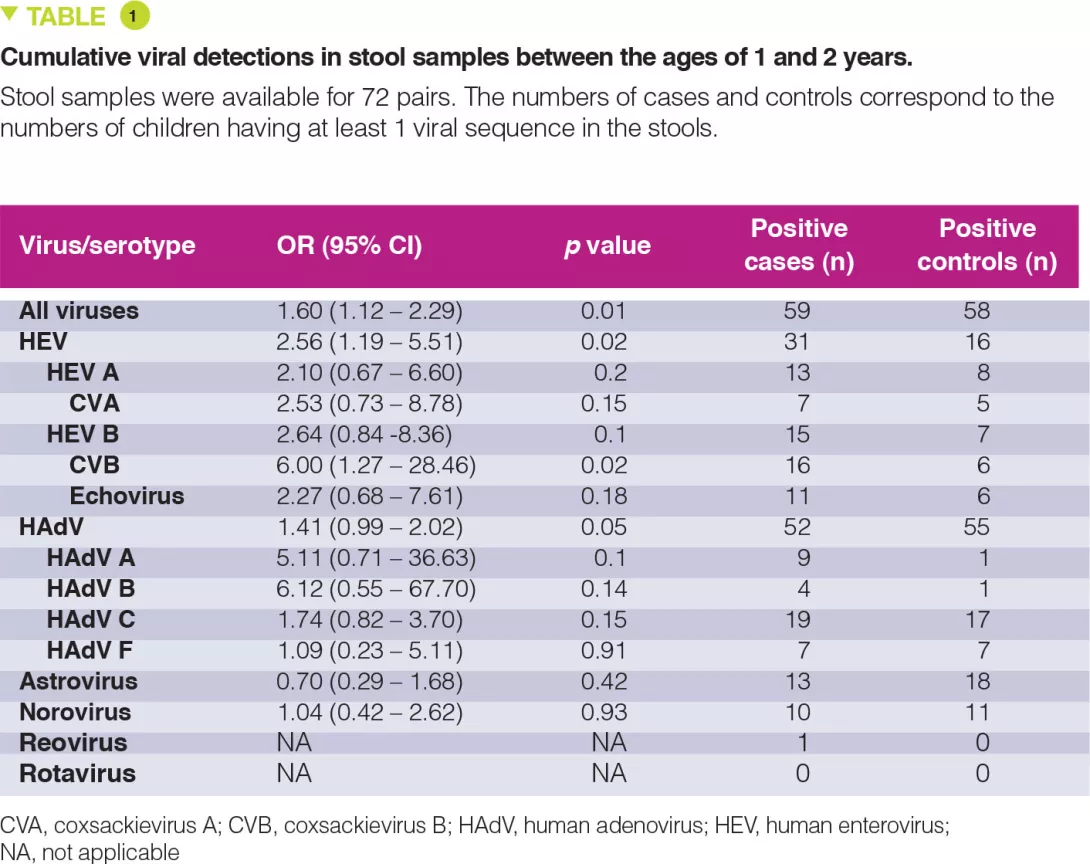
The percentage of stool samples positive for any virus fluctuated from 22 to 50%, without any age-related peak and for the enteroviruses this ranged from 0 to 21% after 6 months. Between 1 and 2 years, enteroviruses were detected in 31 cases versus 16 controls (Table 1). The cumulative number of stool samples positive for any virus was associated with an increased risk for coeliac disease autoimmunity (OR 1.60; p = 0.01), with a stronger association conferred by the enteroviruses (OR 2.56; p = 0.02).
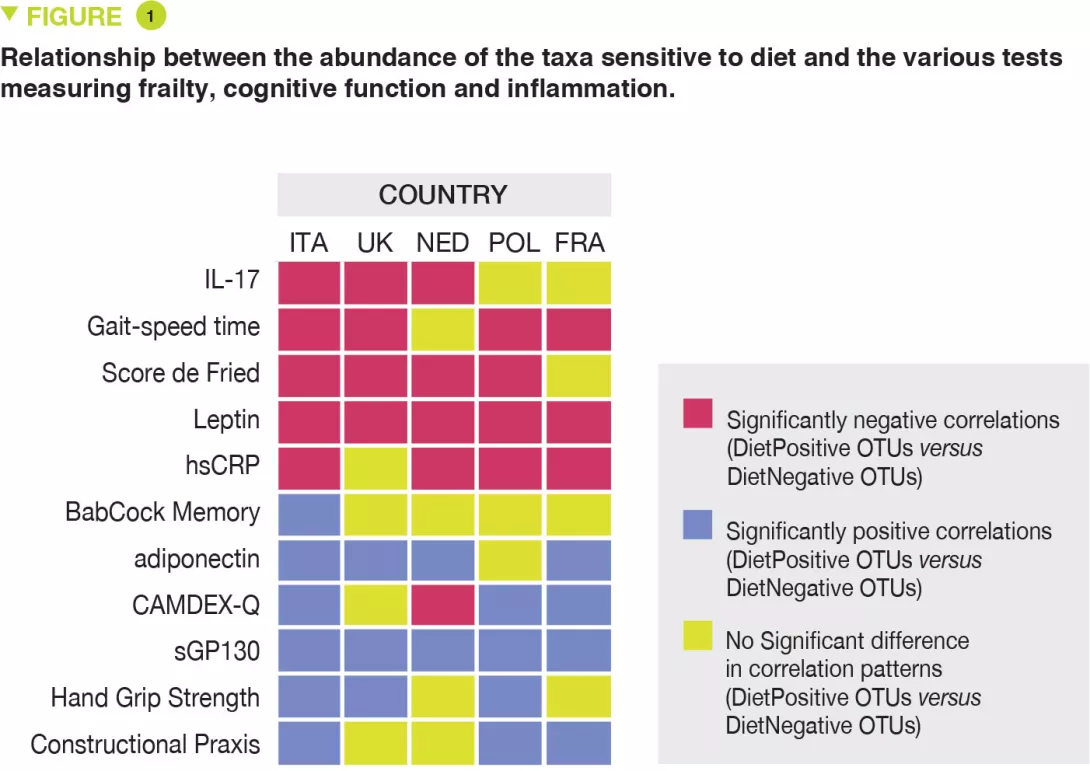
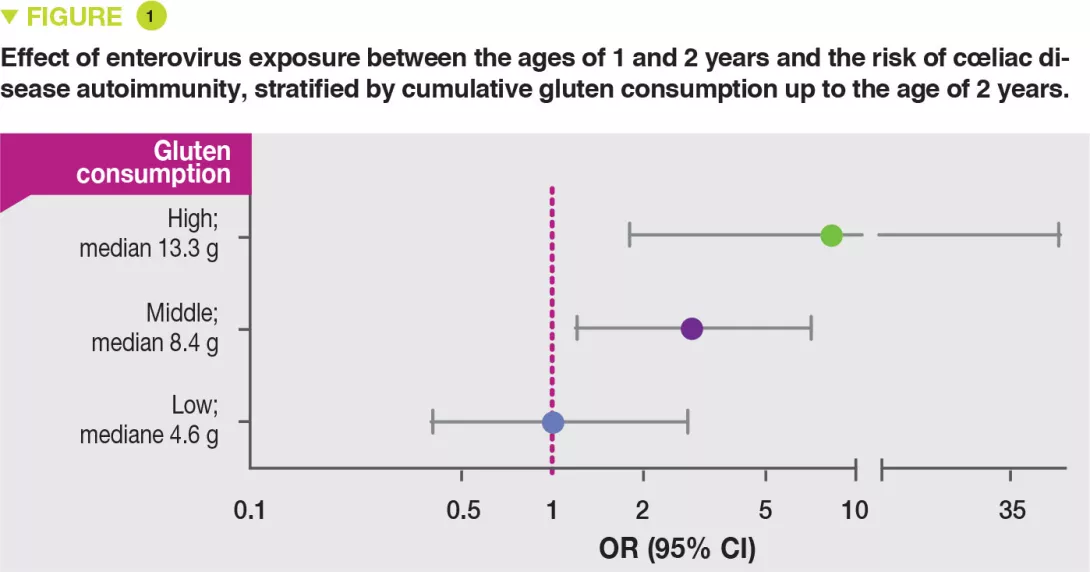
The risk of coeliac disease autoimmunity was not increased by viral infections occurring after the age of gluten introduction while breast-feeding was still continuing. In contrast, after weaning, in stool samples collected between the ages of 1 and 2 years after gluten introduction, both the cumulative number of viruses detected (OR 1.41; p = 0.05) and also the numbers of enteroviruses (OR 2.47; p = 0.03) were associated with the risk of coeliac disease autoimmunity. There was a significant interaction between the presence of enterovirus detected between 1 and 2 years and the quantity of gluten ingested up to the age of 2 years in the risk of coeliac disease autoimmunity (p = 0.03). It is suggested that this increases with the amounts of gluten ingested: high (OR 8.3), middle (OR 2.9) and low (OR 1.0) (Figure 1).
Key points
-
Environmental factors play a role in cœliac disease.
-
Enterovirus exposure is a risk factor for the development of anti-transglutaminase antibodies in HLA DQ2 and/or DQ8-positive children.
-
This risk is potentiated by the intake of large amounts of gluten in the diet.
What are the consequences in practice?
The results of this study indicate the value of preventing the appearance of coeliac disease auto-antibodies in children at risk. This could be done by carefully monitoring the amounts of gluten ingested, in particular in case of exposure to enteroviruses, especially when the child is no longer being breast fed.
Conclusion
This study demonstrated an association between gastro-intestinal exposure to enteroviruses and the risk of coeliac disease autoimmunity in genetically at-risk children. This risk increases as greater amounts of gluten are ingested.